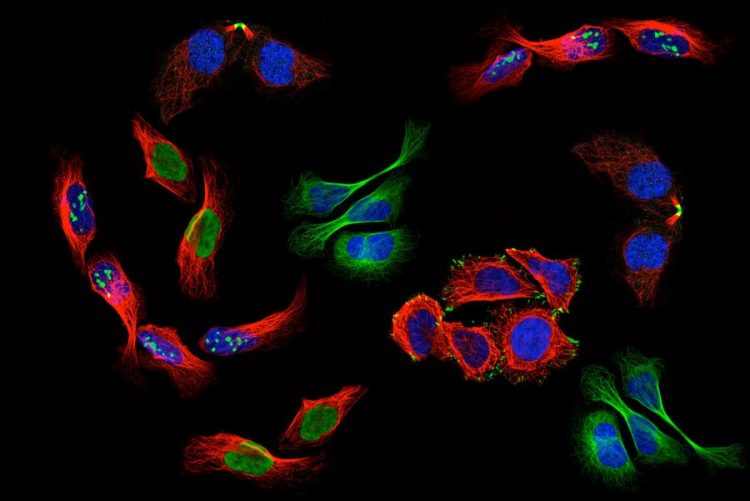
There are billions of humans on this earth, and each of us is made up of trillions of cells. Just like every individual is unique, even genetically identical twins, scientists observe differences between the genetically identical cells in our bodies.
Differences in the location of proteins can give rise to such cellular heterogeneity. Proteins play essential roles in virtually all cellular processes. Often, many different proteins come together at a specific location to perform a task, and the exact outcome of this task depends on which proteins are present. As you can imagine, different subcellular distributions of one protein can give rise to great functional heterogeneity between cells. Finding such differences, and figuring out how and why they occur, is important for understanding how cells function, how diseases develop, and ultimately how to develop better treatments for those diseases.
To see more, start with less. That may seem counterintuitive, but the study of a single cell enables the discovery of mechanisms too difficult to see with multi-cell research. The importance of studying single cells is reflected in the ongoing revolution in biology centered around technologies for single cell analysis. Microscopy offers an opportunity to study differences in protein localizations within a population of cells. Current machine learning models for classifying protein localization patterns in microscope images gives a summary of the entire population of cells. However, the single-cell revolution in biology demands models that can precisely classify patterns in each individual cell in the image.
The Human Protein Atlas is an initiative based in Sweden that is aimed at mapping proteins in all human cells, tissues, and organs. The data in the Human Protein Atlas database is freely accessible to scientists all around the world that allows them to explore the cellular makeup of the human body. Solving the single-cell image classification challenge will help us characterize single-cell heterogeneity in our large collection of images by generating more accurate annotations of the subcellular localizations for thousands of human proteins in individual cells. Thanks to you, we will be able to more accurately model the spatial organization of the human cell and provide new open-access cellular data to the scientific community, which may accelerate our growing understanding of how human cells functions and how diseases develop.
This is a weakly supervised multi-label classification problem and a code competition. Given images of cells from our microscopes and labels of protein location assigned together for all cells in the image, Kagglers will develop models capable of segmenting and classifying each individual cell with precise labels. If successful, you’ll contribute to the revolution of single-cell biology!
The scientific journal Nature Methods is interested in considering a paper discussing the outcome and approaches of the challenge. The Human Protein Atlas team, led by Professor Emma Lundberg, would like to invite top performing teams to join as co-authors in writing this paper. Please follow the discussion forum for more details on how you can help.
Awards:-
- 1st Place – $ 12,000
- 2nd Place – $ 8,000
- 3rd Place – $ 5,000
Deadline:- 04-05-2021